
informatik



<< back | next >>
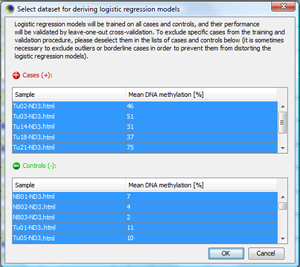
Generate Optimized Biomarker ModelsNow, the optimization step follows, in which single CpG sites in the biomarkers get weights according to their "predictiveness". This is done with logistic regression models. Click on "Train Classifier..." to start the process for checked biomarkers. MethMarker will now ask you for the training set used to train the classifiers. As default, MethMarker uses all loaded samples for training. If you want to exclude a specific sample, just deselect it. For this tutorial, accept all samples and press "OK". |
|
|
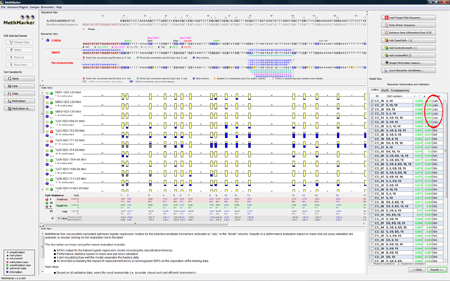
MethMarker has now created logistic regression models for checked biomarkers. This is indicated by a "calc." in the last column of the biomarker table. Biomarkers without regression models are labeled with "N/A". |
|
|
Open the biomarker result window with click on "Results >>". TIP: Alternatively, you can doubleclick on a biomarker with calculated logistic regression model in the biomarker table. |
|
|
Now, you see the
|
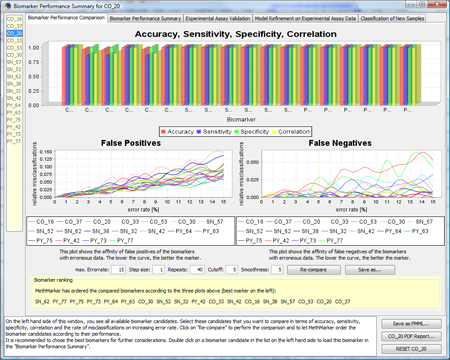
|
|
To get a more detailed overview about the biomarker, click on
If you click on "Error Test", you can see how robustly the model behaves in the presence of erroneous data. For more information about the Error Test, look at the F.A.Q.. TIP: If you don't know how to interpret the regression formula, look at the F.A.Q.. TIP: If you don't know how to interpret accuracy, specificity, sensitivity or correlation, look at the F.A.Q.. |
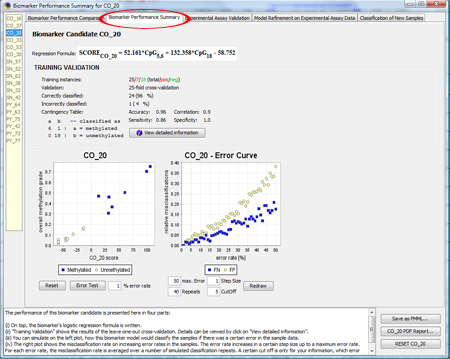
|
|
You can save the model as PMML file (press "Save CO_20 as..." on the right). A PMML file contains the model in a standardized statistical markup language, defined by the data mining group (http://www.dmg.org). Several statistical programms support PMML, and so does MethMarker. For a comprehensive report of this biomarker candidate, click on "CO_20 PDF Report...". The PDF report is for your documents or can be shared with colleagues. MethMarker has now created biomarkers, optimized with logistic regression model and validated with Leave-One-Out Cross-Validation. After saving the models (or saving the whole analysis with "File -> Save Analysis..."), you can close MethMarker. |
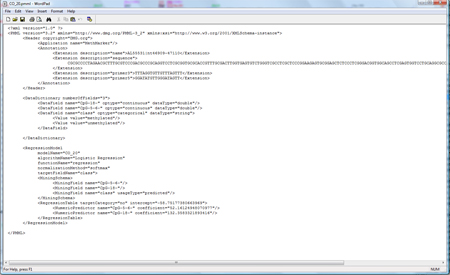
|
<< back | next >> |