
informatik



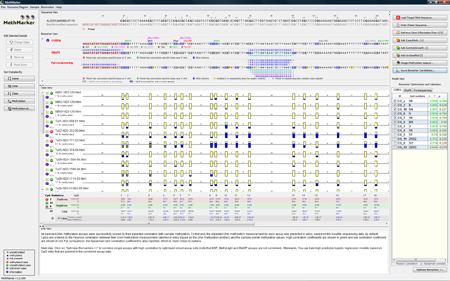
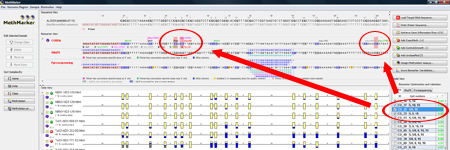
Generate biomarkersClick on "Score Biomarker Candidates...". MethMarker will now assume that each blue highlighted CpG site is a single biomarker. If more than one CpG site lie in one restriction site for Cobra or on one primer for MSP, all these CpGs are handled as a single biomarker. These biomarkers are methylated or not in the different samples. MethMarker correlates the CpG sites with the overall methylation grade of all samples. For more information about the correlation, look at the F.A.Q.. All single CpG sites are listed in the "Model View" in a table. The best correlating biomarkers are checked with a small check mark on the left in the biomarker table. You can check or uncheck biomarkers, if you are interested in models with specific CpG sites. Checked biomarkers will be included in the next step. |
|
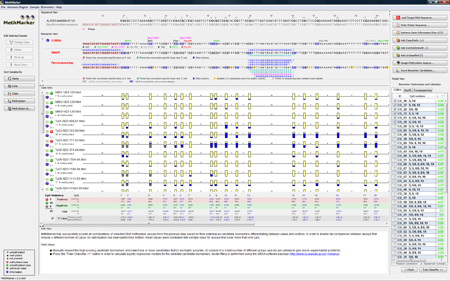
Combine biomarkersClick on "Optimize Biomarkers...". MethMarker will now combine all checked biomarkers with each other. Perhaps, a combination of several CpG sites has a better correlation than the single CpG sites. All combinations are again listed as biomarkers in the "Model View" and again, the best correlating biomarkers are checked to be used in the next step. TIP: Holding the mouse over a table row (biomarker) will highlight the used CpG sites in the "Biomarker View" in red. |
|