
informatik



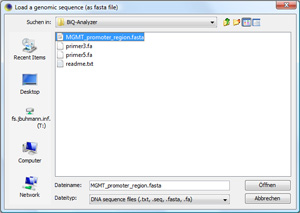
Load genomic target regionClick on "Load Target DNA Sequence..." to load the main sequence, which the biomarker analysis is based on. In our case, this is the MGMT promoter region. Browse through the demonstration data set and open the file |
|
|
MethMarker will open and display the region in the "Sequence View". |

|
|
The MGMT promoter region can be obtained with specific primers. To add these primers, click on "Add Primers" in the Genomic Region menu. TIP: Primers should be added to improve the analysis of marginal CpG sites in the sequence. |
|
|
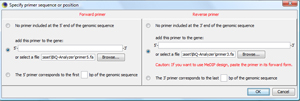
In the primer dialog showing up, select the two primer sequences of the demonstration data set TIP: You can also type in primer sequences directly or determine primer sequences that are already in the loaded target region. |
|
|
Finally, the primers are shown in red. |
|