
informatik



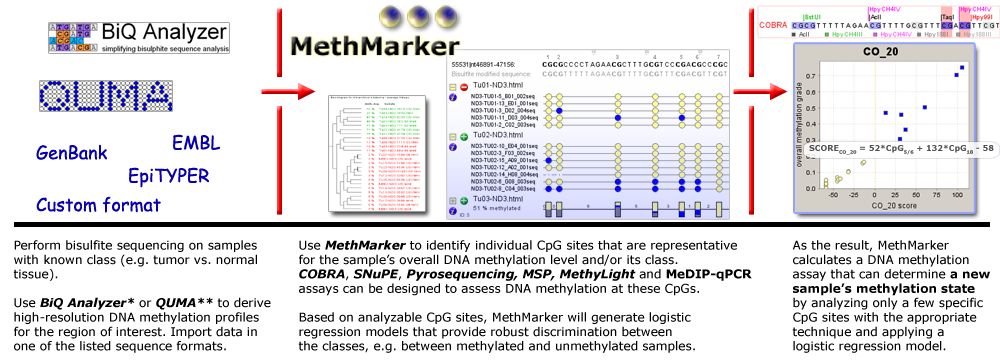
MethMarker facilitates the design and optimization of gene-specific DNA methylation assays. Beyond its use as an epigenetic primer-design tool, it provides extensive support for epigenetic biomarker optimization. For a given gene or genomic region, MethMarker helps design DNA methylation assays for any of the following protocols:
Furthermore, if DNA methylation profiles are available for the region of interest, MethMarker can be used to optimize and validate DNA methylation biomarkers that provide robust classification, for example between tumor samples and healthy control tissue. If high-resolution DNA methylation profiles are available for the region of interest (e.g. from bisulfite sequencing, http://biq-analyzer.bioinf.mpi-inf.mpg.de), MethMarker can exploit these data to design biomarker assays that optimally discriminate between cases (e.g. cancer samples) and controls. |
|
* http://biq-analyzer.bioinf.mpi-inf.mpg.de
** http://quma.cdb.riken.jp/
Click here to download MethMarker or to start it directly from within your web browser.
Click here to see documentation material on MethMarker as well as a step-by-step tutorial.
Peter Schüffler, Christoph Bock, Thomas Mikeska and Thomas Lengauer
Please cite MethMarker as you use it for your work:
MethMarker: User-friendly design and optimization of gene-specific DNA methylation assays
Peter Schüffler, Thomas Mikeska, Andreas Waha, Thomas Lengauer, Christoph Bock
Genome Biology 2009, 10:R105doi:10.1186/gb-2009-10-10-r105
If you have any questions, comments, suggestions or criticism, feel free to contact us at methmarker@mpi-inf.mpg.de.
MethMarker comes "as is", with no warranty with respect to its fitness for purpose, its operational state, character, quality, or freedom from defects, or the non-infringement of rights of third parties.